3D printing is useful in how it makes tools for us humans. Tools are the gateway to innovation within our world. 3D Printing helps different industries because of cost reductions and material waste. We are going to look into a specific industry application and how it is leveraging 3D printing technology.
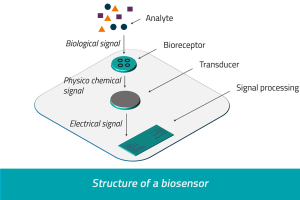
A biosensor is an analytical device, used for the detection of a chemical substance, that combines a biological component with a physicochemical detector. The sensitive biological element, e.g. tissue, microorganisms, organelles, cell receptors, enzymes, antibodies, nucleic acids, etc., is a biologically derived material or biomimetic component that interacts, binds, or recognizes with the analyte under study.
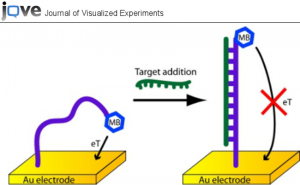
We are particularly interested in DNA biosensors. DNA biosensors can theoretically be used for medical diagnostics, forensic science, agriculture, or even environmental clean-up efforts. No external monitoring is needed for DNA-based sensing devices. Typically these machines were very large and expensive devices that were only for research purposes. DNA biosensors are now becoming complicated mini-machines—consisting of sensing elements, micro lasers, and a signal generator. This means that DIY construction of such a device will lead to better public health implications for makers. DNA biosensors function on the fact that two strands of DNA stick to each other through chemical attractive forces. Only an exact fit—that is, two strands that match up at every nucleotide position leads to a fluorescent signal that is then transmitted to a signal generator.
The field of biosensors has developed extensively and now has become one of the essential state of the art technologies in laboratory medicine. The idea of biosensors has revolutionized the concept of self-testing by the patient in many clinical conditions. Quick diagnosis and early prevention are critical for the control of disease status. Some commercial biosensors on the market include the following:
So why are 3D printers important in the field of biosensing? In terms of tool creation, it is vital for biotech companies and researchers to build new prototypes and mini machines for biosensing. In order to rapidly make advances within this type of technology, 3D printing is used to cut down costs, as well as iterate designs. This helps to streamline innovation and develop new methods to reduce the production of biosensors.
Researchers currently are working on the potential of adopting 3D printing technology for electrochemical DNA biosensing applications. Some groups have created helical-shaped stainless steel electrodes that are vital for biosensing. An electrode is an electrical conductor used to make contact with a nonmetallic part of a circuit (e.g. a semiconductor, an electrolyte, a vacuum or air). This connection is vital for signal processing and seeing whether or not DNA can be identified within a biosensor.
These can be designed and 3D printed through the use of a selective laser melting (SLM) method, which fuses a fine metal powder on a printing stage with a high intensity laser beam, in a layer-by-layer manner. SLM is also called Laser Powder Bed Fusion and is similar to DMLS or Direct Metal Laser Sintering while commonly being called metal 3D printing. It is important to use metal printing for a biosensor because specific chemical and electrical reactions occur with the use of different metal groups.
The future of biotech is oriented towards how devices can be made quickly and cheaply. With 3D printing, a maker who is curious has a lot of tools and the ability to make devices efficiently. This leads to projects and innovations when one is open to iterating and experimenting. More importantly one should learn how to create their own devices. Health care as a whole is leaning toward preventative measures. It is important to have tools that can diagnose complications that we are not aware of. It also is important due to the fact that healthcare will be more expensive as time continues. For underdeveloped nations, having access to open source solutions to biotechnology projects can lead to better maintenance of their citizens and their general health.
The future of health is reliant on different technologies, and 3D printing is a very essential part of the equation. With 3D printing, we are able to create tools that were previously only accessible to industry experts. Now a common consumer with an interest can create medical tools such as a biosensor to assist them in their own general health. What will we as makers do to help that?